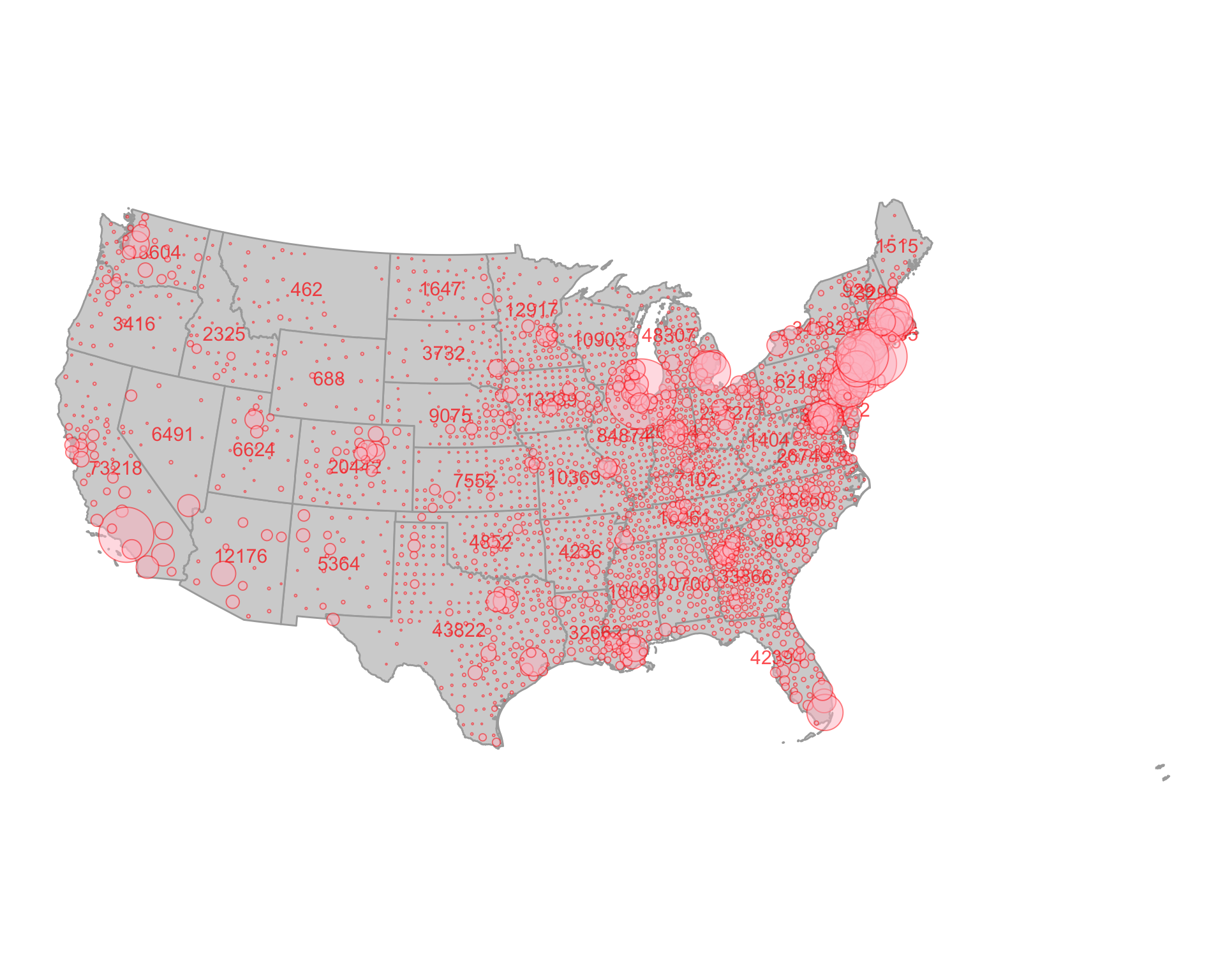
The NY Times has been making an amazing infographic of Covid-19 cases. With the data from this package, we should be able to more or less recreate it. Let’s do it for the lower 48 (moving Hawaii and Alaska is a pain!)
We’ll start by loading the coronavirus data at the county and state level
require(covid19nytimes) require(dplyr) covid19nytimes_states <- refresh_covid19nytimes_states() %>% filter(date == max(date)) covid19nytimes_counties <- refresh_covid19nytimes_counties()%>% filter(date == max(date))
Next, we need some county maps and state maps. For this, we’ll use the excellent tigris package. We want the centroid of counties for bubble placement.
require(sf) require(tigris) county_map <- counties(cb = TRUE, resolution = '20m', class = "sf") %>% st_centroid() state_map <- states(cb = TRUE, resolution = '5m', class = "sf") #make a state lookup table state_lookup <- state_map %>% as_tibble() %>% select(STATEFP, NAME) %>% rename(STATE_NAME = NAME)
OK, let’s filter to the lower 48 for all data sets
filter_out <- c("Alaska", "Hawaii", "Guam", "Puerto Rico", "American Samoa", "Commonwealth of the Northern Mariana Islands", "Virgin Islands") covid19nytimes_states <- covid19nytimes_states %>% filter(!(location %in% filter_out)) %>% filter(location != "Unknown") covid19nytimes_counties <- covid19nytimes_counties %>% filter(purrr::map_dbl(location, ~stringr::str_detect(., filter_out) %>% sum)==0) %>% filter(location != "Unknown") county_map <- county_map %>% left_join(state_lookup) %>% filter(!(STATE_NAME %in% filter_out)) #> Joining, by = "STATEFP" state_map <- state_map %>% filter(!(NAME %in% filter_out))
For text labels, we use centroids of states joined with maximum number of case numbers.
state_num_centroids <- covid19nytimes_states %>% filter(date == max(date)) %>% filter(data_type == "cases_total") %>% left_join(state_map, by = c("location_code" = "STATEFP")) %>% st_as_sf(crs = st_crs(state_map)) %>% st_centroid()
Now, we join the county data and the county centroid map and filter to the last date in the data set.
joined_covid_counties <- covid19nytimes_counties %>% filter(data_type == "cases_total") %>% left_join(county_map, by = c("location_code" = "GEOID")) %>% st_as_sf(crs = st_crs(county_map))
Last, let’s make the plot!
require(ggplot2) #> Loading required package: ggplot2 ggplot() + geom_sf(data = state_map, fill = "lightgrey", color = "darkgrey") + geom_sf_text(data = state_num_centroids, aes(label = value), color = "red", size = 4, alpha = 0.7) + geom_sf(data = joined_covid_counties, mapping = aes(size = value), shape = 21, alpha = 0.5, color = "red", fill = "pink") + theme_void() + scale_size_continuous(guide= "none", range = c(0.05, 35)) + coord_sf(crs = st_crs(2163)) #for that nice equal area curved look